2023-10-19 ミュンヘン大学(LMU)
◆このソフトウェアは、データを自動的に評価し、信頼性を向上させ、迅速に情報を抽出します。 Deep-LASIは深層学習を活用し、ニューラルネットワークを使用してデータの解析を自動化し、効率的に行います。
◆その結果、非常に高い信頼性と精度で作業し、競合ソフトウェアや人間を上回ります。ニューラルネットワークはデータ内の複雑なパターンを認識し、人間よりも迅速に分析でき、偏りが少ないため、精度が向上します。
<関連情報>
- https://www.lmu.de/en/newsroom/news-overview/news/using-ai-to-understand-the-nano-world.html
- https://www.nature.com/articles/s41467-023-42272-9
Deep-LASI:深層学習支援によるマルチカラーDNA折り紙構造の単一分子イメージング解析 Deep-LASI: deep-learning assisted, single-molecule imaging analysis of multi-color DNA origami structures
Simon Wanninger,Pooyeh Asadiatouei,Johann Bohlen,Clemens-Bässem Salem,Philip Tinnefeld,Evelyn Ploetz & Don C. Lamb
Nature Communications Published:17 October 2023
DOI:https://doi.org/10.1038/s41467-023-42272-9
Abstract
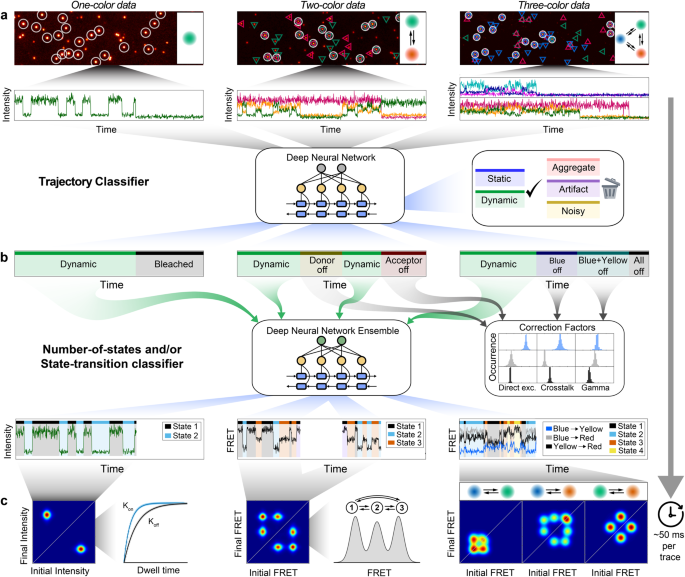
Single-molecule experiments have changed the way we explore the physical world, yet data analysis remains time-consuming and prone to human bias. Here, we introduce Deep-LASI (Deep-Learning Assisted Single-molecule Imaging analysis), a software suite powered by deep neural networks to rapidly analyze single-, two- and three-color single-molecule data, especially from single-molecule Förster Resonance Energy Transfer (smFRET) experiments. Deep-LASI automatically sorts recorded traces, determines FRET correction factors and classifies the state transitions of dynamic traces all in ~20–100 ms per trajectory. We benchmarked Deep-LASI using ground truth simulations as well as experimental data analyzed manually by an expert user and compared the results with a conventional Hidden Markov Model analysis. We illustrate the capabilities of the technique using a highly tunable L-shaped DNA origami structure and use Deep-LASI to perform titrations, analyze protein conformational dynamics and demonstrate its versatility for analyzing both total internal reflection fluorescence microscopy and confocal smFRET data.