2024-08-12 ペンシルベニア州立大学(PennState)
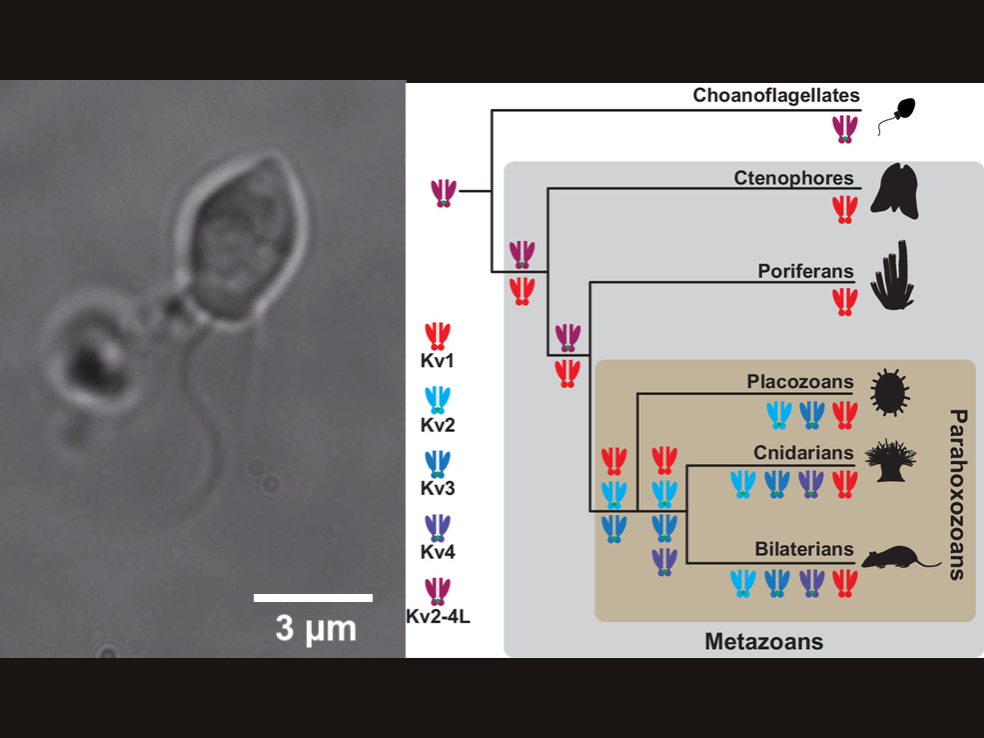 Researchers have found certain ion channels, proteins critical for electrical signaling in the nervous system, in single celled-organisms called choanoflagellates that are the closest living relatives to animals. This rewrites the evolutionary tree of these ion channels, which were previously thought to be only be found in animals. Left: Choanoflagellate under the microscope. Right: Proposed evolutionary tree of the various types of Shaker family ion channels in choanoflagellates, top, and animals, or Metazoans. Credit: Image: David Spafford; Tree: Jegla et al. 2024, PNAS. All Rights Reserved.
Researchers have found certain ion channels, proteins critical for electrical signaling in the nervous system, in single celled-organisms called choanoflagellates that are the closest living relatives to animals. This rewrites the evolutionary tree of these ion channels, which were previously thought to be only be found in animals. Left: Choanoflagellate under the microscope. Right: Proposed evolutionary tree of the various types of Shaker family ion channels in choanoflagellates, top, and animals, or Metazoans. Credit: Image: David Spafford; Tree: Jegla et al. 2024, PNAS. All Rights Reserved.
<関連情報>
- https://www.psu.edu/news/eberly-college-science/story/rewriting-evolutionary-history-critical-components-nervous-system/
- https://www.pnas.org/doi/10.1073/pnas.2407461121
襟鞭毛虫の広範な調査から、動物における電位依存性K+チャネルのシェーカー・ファミリーの進化の歴史が見直される A broad survey of choanoflagellates revises the evolutionary history of the Shaker family of voltage-gated K+ channels in animals
Timothy Jegla, Benjamin T. , and J. David Spafford
Proceedings of the National Academy of Sciences Published:July 17, 2024
DOI:https://doi.org/10.1073/pnas.2407461121
Significance
Shaker family voltage-gated K+ channels are arguably the most studied ion channels, having long served as workhorses for ion channel biophysics, structural biology, and molecular genetic dissection of neuronal excitability. Members of the gene family encode many of the canonical neuronal K+ currents, including classical delayed rectifiers first described by Hodgkin and Huxley and a variety of inactivating A-type currents. Here, we show that the Shaker family must have already been present in the protozoan ancestor of choanoflagellates and animals and that stem animals must have had at least two distinct molecular lineages of Shaker family channels. The Choanoflagellate Shaker family genes encode classic A-type channels with rapid N-type ball and chain inactivation; functionally similar channels are widespread in animals.
Abstract
The Shaker family of voltage-gated K+ channels has been thought of as an animal-specific ion channel family that diversified in concert with nervous systems. It comprises four functionally independent gene subfamilies (Kv1-4) that encode diverse neuronal K+ currents. Comparison of animal genomes predicts that only the Kv1 subfamily was present in the animal common ancestor. Here, we show that some choanoflagellates, the closest protozoan sister lineage to animals, also have Shaker family K+ channels. Choanoflagellate Shaker family channels are surprisingly most closely related to the animal Kv2-4 subfamilies which were believed to have evolved only after the divergence of ctenophores and sponges from cnidarians and bilaterians. Structural modeling predicts that the choanoflagellate channels share a T1 Zn2+ binding site with Kv2-4 channels that is absent in Kv1 channels. We functionally expressed three Shakers from Salpingoeca helianthica (SheliKvT1.1-3) in Xenopus oocytes. SheliKvT1.1-3 function only in two heteromultimeric combinations (SheliKvT1.1/1.2 and SheliKvT1.1/1.3) and encode fast N-type inactivating K+ channels with distinct voltage dependence that are most similar to the widespread animal Kv1-encoded A-type Shakers. Structural modeling of the T1 assembly domain supports a preference for heteromeric assembly in a 2:2 stoichiometry. These results push the origin of the Shaker family back into a common ancestor of metazoans and choanoflagellates. They also suggest that the animal common ancestor had at least two distinct molecular lineages of Shaker channels, a Kv1 subfamily lineage predicted from comparison of animal genomes and a Kv2-4 lineage predicted from comparison of animals and choanoflagellates.

