2024-09-11 テキサス大学オースチン校(UT Austin)
<関連情報>
- https://news.utexas.edu/2024/09/11/newly-discovered-antimicrobial-could-prevent-or-treat-cholera/
- https://www.sciencedirect.com/science/article/abs/pii/S1931312824003196?via%3Dihub
- https://www.biorxiv.org/content/10.1101/2023.11.15.567263v1
コレラ菌マイクロシンの抗菌作用、タンパク質分解免疫、およびin vivo活性 Antibacterial action, proteolytic immunity, and in vivo activity of a Vibrio cholerae microcin
Sun-Young Kim, Justin R. Randall, Richard Gu, Quoc D. Nguyen, Bryan W. Davies
Cell Host & Microbe Published: September 10, 2024
DOI:https://doi.org/10.1016/j.chom.2024.08.012
Graphical abstract

Highlights
- Vibrio cholerae encodes microcin systems
- Microcin MvcC kills virulent V. cholerae strains in vitro and in vivo
- MvcC uses OmpT and OppA to reach and permeabilize the inner membrane
- MvcD is a microcin immunity protein that neutralizes MvcC by endoproteolysis
Summary
Microcins are small antibacterial proteins that mediate interbacterial competition. Their narrow-spectrum activity provides opportunities to discover microbiome-sparing treatments. However, microcins have been found almost exclusively in Enterobacteriaceae. Their broader existence and potential implications in other pathogens remain unclear. Here, we identify and characterize a microcin active against pathogenic Vibrio cholerae: MvcC. We show that MvcC is reliant on the outer membrane porin OmpT to cross the outer membrane. MvcC then binds the periplasmic protein OppA to reach and disrupt the cytoplasmic membrane. We demonstrate that MvcC’s cognate immunity protein is a protease, which precisely cleaves MvcC to neutralize its activity. Importantly, we show that MvcC is active against diverse cholera isolates and in a mouse model of V. cholerae colonization. Our results provide a detailed analysis of a microcin outside of Enterobacteriaceae and its potential to influence V. cholerae infection.
タンパク質大規模言語モデルを用いた意味検索により細菌ゲノム中のクラスIIマイクロシンが検出される Semantic search using protein large language models detects class II microcins in bacterial genomes
Anastasiya V. Kulikova, Jennifer K. Parker, Bryan W. Davies, Claus O. Wilke
bioRxiv Posted: November 15, 2023
DOI:https://doi.org/10.1101/2023.11.15.567263
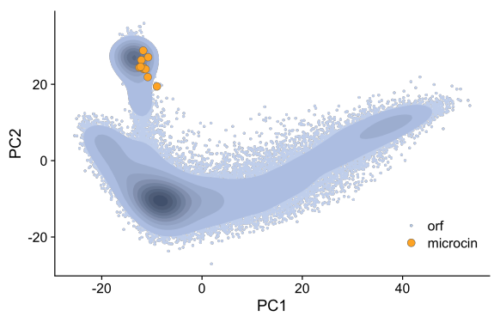
Abstract
Class II microcins are antimicrobial peptides that have shown some potential as novel antibiotics. However, to date only ten class II microcins have been described, and discovery of novel microcins has been hampered by their short length and high sequence divergence. Here, we ask if we can use numerical embeddings generated by protein large language models to detect microcins in bacterial genome assemblies and whether this method can outperform sequence-based methods such as BLAST. We find that embeddings detect known class II microcins much more reliably than does BLAST and that any two microcins tend to have a small distance in embedding space even though they typically are highly diverged at the sequence level. In datasets of Escherichia coli, Klebsiella spp., and Enterobacter spp. genomes, we further find novel putative microcins that were previously missed by sequence-based search methods.