2025-05-09 東京大学
<関連情報>
- https://www.u-tokyo.ac.jp/content/400263769.pdf
- https://genomebiology.biomedcentral.com/articles/10.1186/s13059-025-03583-w
ヒトB細胞における新規アイソフォーム特異的eQTLとアイソフォーム発現制御機構がロングリードシークエンシングにより明らかになった Long-read sequencing reveals novel isoform-specific eQTLs and regulatory mechanisms of isoform expression in human B cells
Yuya Nagura,Mihoko Shimada,Ryoji Kuribayashi,Ko Ikemoto,Hiroki Kiyose,Arisa Igarashi,Tadashi Kaname,Motoko Unoki & Akihiro Fujimoto
Genome Biology Published:08 May 2025
DOI:https://doi.org/10.1186/s13059-025-03583-w
Abstract
Background
Genetic variations linked to changes in gene expression are known as expression quantitative loci (eQTLs). The identification of eQTLs helps to understand the mechanisms governing gene expression. However, prior studies have primarily utilized short-read sequencing techniques, and the analysis of eQTLs on isoforms has been relatively limited.
Results
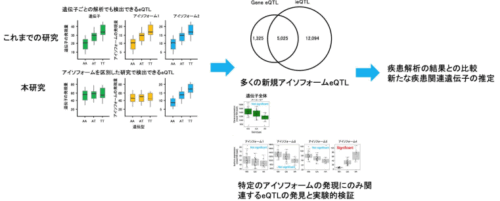
In this study, we employ long-read sequencing technology (Oxford Nanopore) on B cells from 67 healthy Japanese individuals to explore genetic variations associated with isoform expression levels, referred to as isoform eQTLs (ieQTLs). Our analysis reveals 17,119 ieQTLs, with 70.6% remaining undetected by a gene-level analysis. Additionally, we identify ieQTLs that have significantly different effects on isoform expression levels within a gene. A functional feature analysis demonstrates a significant enrichment of ieQTLs at splice sites and specific histone marks, such as H3K36me3, H3K4me1, H3K4me3, and H3K79me2. Through an experimental validation using genome editing, we observe that a distant genomic region can modulate isoform-specific expression. Moreover, an ieQTL analysis and minigene splicing assays unveils functionally crucial variants in splicing that splicing prediction software did not assign a high prediction score. A comparison with GWAS data reveals a higher number of colocalizations between ieQTLs and GWAS findings compared to gene eQTLs.
Conclusions
These findings highlight the substantial contribution of ieQTLs identified through long-read analysis in our understanding of the functional implications of genetic variations and the regulatory mechanisms governing isoforms.