2025-06-04 カリフォルニア大学サンディエゴ校(UCSD)
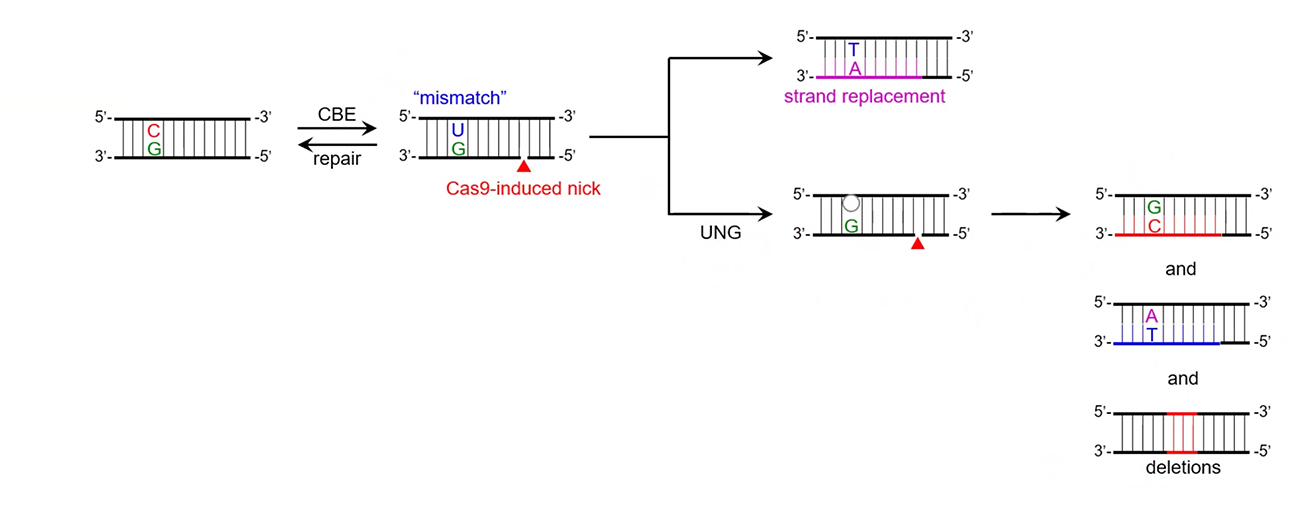 A schematic showing the cytosine base editing intermediate and the various outcomes that can occur when the UNG protein is active.
A schematic showing the cytosine base editing intermediate and the various outcomes that can occur when the UNG protein is active.
<関連情報>
- https://today.ucsd.edu/story/how-gene-editing-tools-work
- https://www.nature.com/articles/s41467-025-59948-z
CRISPRiスクリーニングによるシトシン塩基編集の成果を支配する遺伝的メカニズムの解明 Elucidating the genetic mechanisms governing cytosine base editing outcomes through CRISPRi screens
Sifeng Gu,Zsolt Bodai,Rachel A. Anderson,Hei Yu Annika So,Quinn T. Cowan & Alexis C. Komor
Nature Communications Published:20 May 2025
DOI:https://doi.org/10.1038/s41467-025-59948-z
Abstract
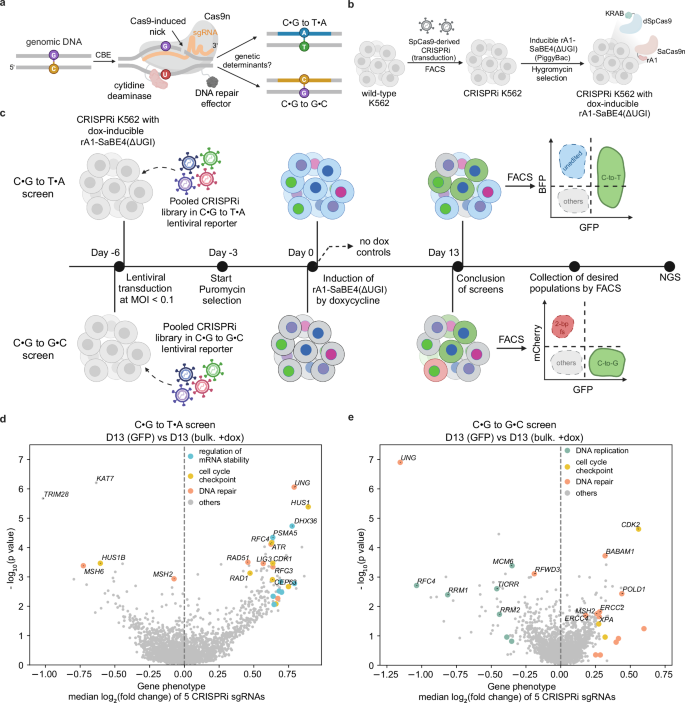
Cytosine base editors enable programmable and efficient genome editing using an intermediate featuring a U•G mismatch across from a DNA nick. This intermediate facilitates two major outcomes, C•G to T•A and C•G to G•C point mutations, and it is not currently well-understood which DNA repair factors are involved. Here, we couple reporters for cytosine base editing activity with knockdown of 2015 DNA processing genes to identify genes involved in these two outcomes. Our data suggest that mismatch repair factors facilitate C•G to T•A outcomes, while C•G to G•C outcomes are mediated by RFWD3, an E3 ubiquitin ligase. We also propose that XPF, a 3’-flap endonuclease, and LIG3, a DNA ligase, are involved in repairing the intermediate back to the original C•G base pair. Our results demonstrate that competition and collaboration among different DNA repair pathways shape cytosine base editing outcomes.

