2025-09-11 ウースター工科大学(WPI)
<関連情報>
- https://www.wpi.edu/news/announcements/wpi-researchers-design-microbial-tool-analyze-neuropeptide-function-advance-may-point-new-approach
- https://academic.oup.com/genetics/advance-article-abstract/doi/10.1093/genetics/iyaf155/8223085
微生物ツールの活用:線虫(Caenorhabditis elegans)における神経ペプチド機能解析のための宿主としての大腸菌(Escherichia coli) Harnessing microbial tools: Escherichia coli as a vehicle for neuropeptide functional analysis in Caenorhabditis elegans
Elizabeth M DiLoreto, Shruti Shastry, Emily J Leptich, Douglas K Reilly, Rachel N Arey, Jagan Srinivasan
Genetics Published:06 August 2025
DOI:https://doi.org/10.1093/genetics/iyaf155
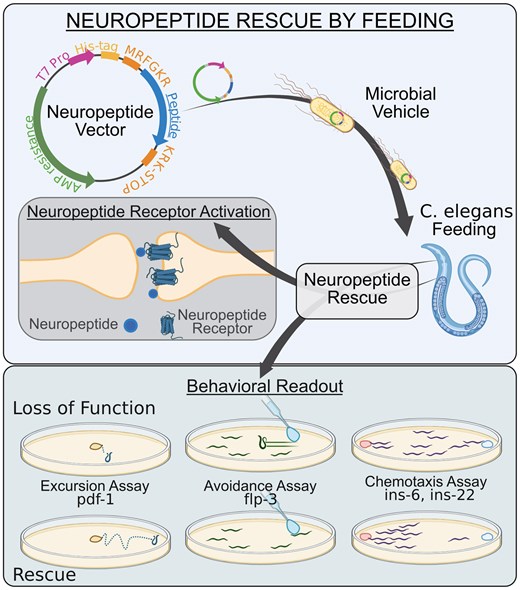
Graphical Abstract
Abstract
Animals respond to changes in their environment and internal states via neuromodulation. Neuropeptides modulate neural circuits with flexibility because 1 gene can produce either multiple copies of the same neuropeptide or different neuropeptides. However, with this architectural complexity, the function of discrete and active neuropeptides is muddled. Here, we design a genetic tool that facilitates functional analysis of individual peptides. We engineered Escherichia coli bacteria to express active peptides, fed loss-of-function Caenorhabditis elegans, and rescued the activity of genes with varying lengths and functions: pdf-1, flp-3, ins-6, and ins-22. Some peptides were functionally redundant, while others exhibit unique and previously uncharacterized functions. We postulate our rescue-by-feeding approach can elucidate the functional landscape of neuropeptides, identifying the circuits and complex peptidergic pathways that regulate different behavioral and physiological processes.

