2025-10-06 ハーバード大学
<関連情報>
- https://seas.harvard.edu/news/2025/10/order-disordered-proteins
- https://www.nature.com/articles/s43588-025-00881-y
本質的に無秩序なタンパク質の配列-アンサンブル-機能関係の一般化設計 Generalized design of sequence–ensemble–function relationships for intrinsically disordered proteins
Ryan K. Krueger,Michael P. Brenner & Krishna Shrinivas
Nature Computational Science Published:06 October 2025
DOI:https://doi.org/10.1038/s43588-025-00881-y
Abstract
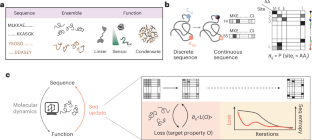
The design of folded proteins has advanced substantially in recent years. However, many proteins and protein regions are intrinsically disordered and lack a stable fold, that is, the sequence of an intrinsically disordered protein (IDP) encodes a vast ensemble of spatial conformations that specify its biological function. This conformational plasticity and heterogeneity makes IDP design challenging. Here we introduce a computational framework for de novo design of IDPs through rational and efficient inversion of molecular simulations that approximate the underlying sequence–ensemble relationship. We highlight the versatility of this approach by designing IDPs with diverse properties and arbitrary sequence constraints. These include IDPs with target ensemble dimensions, loops and linkers, highly sensitive sensors of physicochemical stimuli, and binders to target disordered substrates with distinct conformational biases. Overall, our method provides a general framework for designing sequence–ensemble–function relationships of biological macromolecules.