2022-04-28 オークリッジ国立研究所(ORNL)
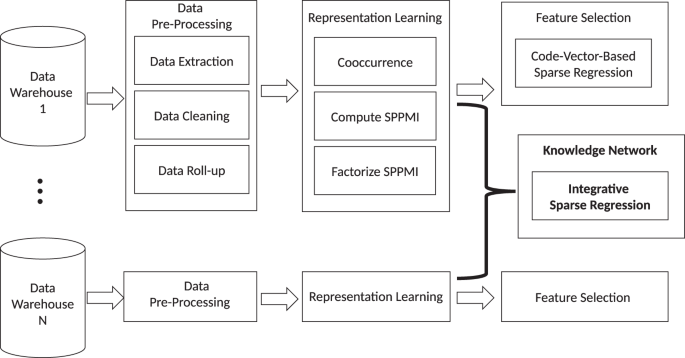
この手法は、Knowledge Extraction via Sparse Embedding Regression(KESER)と呼ばれ、最近Nature Digital Medicineに掲載されました。このプロセスは、米国バージニア州とボストンに拠点を置くパートナーズ・ヘルスケアという2つの大規模施設の電子カルテデータを統合し、表現型識別アルゴリズムと知識発見につながる自動特徴選択を提供するものです。
<関連情報>
- https://www.ornl.gov/news/va-ornl-and-harvard-develop-novel-method-identify-complex-medical-relationships
- https://www.nature.com/articles/s41746-021-00519-z
多施設大規模電子カルテデータを用いたスパース埋め込み回帰による臨床知識抽出(KESER) Clinical knowledge extraction via sparse embedding regression (KESER) with multi-center large scale electronic health record data
Chuan Hong,Everett Rush,Molei Liu,Doudou Zhou,Jiehuan Sun,Aaron Sonabend,Victor M. Castro,Petra Schubert,Vidul A. Panickan,Tianrun Cai,Lauren Costa,Zeling He,Nicholas Link,Ronald Hauser,J. Michael Gaziano,Shawn N. Murphy,George Ostrouchov,Yuk-Lam Ho,Edmon Begoli,Junwei Lu,Kelly Cho,Katherine P. Liao,Tianxi Cai & VA Million Veteran Program
Nature Digital Medicine Published: 27 October 2021
DOI:https://doi.org/10.1038/s41746-021-00519-z
Abstract
The increasing availability of electronic health record (EHR) systems has created enormous potential for translational research. However, it is difficult to know all the relevant codes related to a phenotype due to the large number of codes available. Traditional data mining approaches often require the use of patient-level data, which hinders the ability to share data across institutions. In this project, we demonstrate that multi-center large-scale code embeddings can be used to efficiently identify relevant features related to a disease of interest. We constructed large-scale code embeddings for a wide range of codified concepts from EHRs from two large medical centers. We developed knowledge extraction via sparse embedding regression (KESER) for feature selection and integrative network analysis. We evaluated the quality of the code embeddings and assessed the performance of KESER in feature selection for eight diseases. Besides, we developed an integrated clinical knowledge map combining embedding data from both institutions. The features selected by KESER were comprehensive compared to lists of codified data generated by domain experts. Features identified via KESER resulted in comparable performance to those built upon features selected manually or with patient-level data. The knowledge map created using an integrative analysis identified disease-disease and disease-drug pairs more accurately compared to those identified using single institution data. Analysis of code embeddings via KESER can effectively reveal clinical knowledge and infer relatedness among codified concepts. KESER bypasses the need for patient-level data in individual analyses providing a significant advance in enabling multi-center studies using EHR data.