イネが干ばつや洪水に耐えるための遺伝子的な知見 Genetic insights help rice survive drought and flood
2022-05-19 カリフォルニア大学リバーサイド校(UCR)
<関連情報>
- https://news.ucr.edu/articles/2022/05/19/new-strategies-save-worlds-most-indispensable-grain
- https://www.cell.com/developmental-cell/fulltext/S1534-5807(22)00253-2
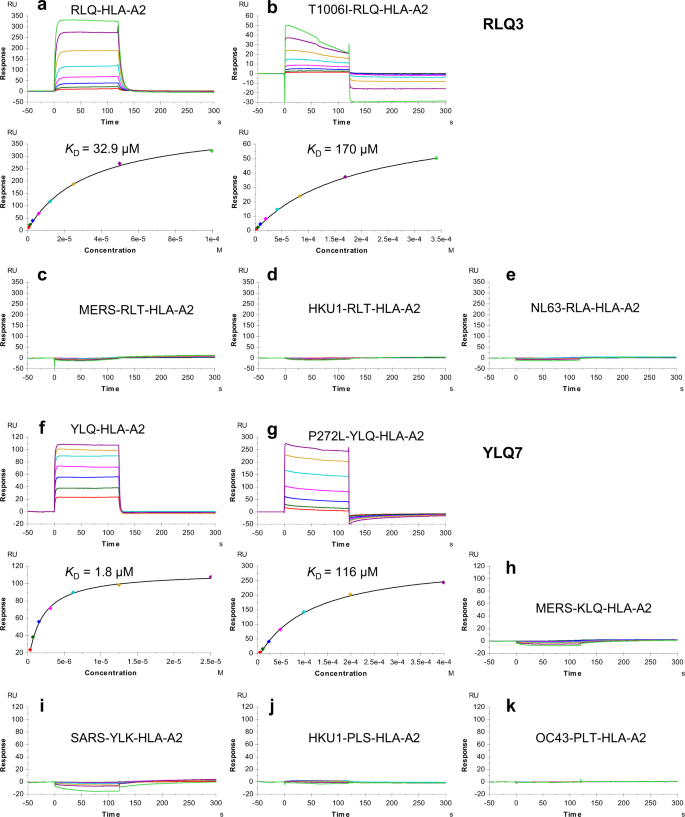
イネの極端な水環境下における根の細胞型の発生的可塑性を形成する遺伝子制御ネットワーク Gene regulatory networks shape developmental plasticity of root cell types under water extremes in rice
Mauricio A. Reynoso,Alexander T. Borowsky,Germain C. Pauluzzi ,Elaine Yeung,Jianhai Zhang,Elide Formentin,Joel Velasco,Sean Cabanlit,Christine Duvenjian,Matthew J. Prior,Garo Z. Akmakjian,Roger B. Deal,Neelima R. Sinha,Siobhan M. Brady,Thomas Girke,Julia Bailey-Serres
Developmental Cell Published:May 02, 2022
DOI:https://doi.org/10.1016/j.devcel.2022.04.013

Summary
Understanding how roots modulate development under varied irrigation or rainfall is crucial for development of climate-resilient crops. We established a toolbox of tagged rice lines to profile translating mRNAs and chromatin accessibility within specific cell populations. We used these to study roots in a range of environments: plates in the lab, controlled greenhouse stress and recovery conditions, and outdoors in a paddy. Integration of chromatin and mRNA data resolves regulatory networks of the following: cycle genes in proliferating cells that attenuate DNA synthesis under submergence; genes involved in auxin signaling, the circadian clock, and small RNA regulation in ground tissue; and suberin biosynthesis, iron transporters, and nitrogen assimilation in endodermal/exodermal cells modulated with water availability. By applying a systems approach, we identify known and candidate driver transcription factors of water-deficit responses and xylem development plasticity. Collectively, this resource will facilitate genetic improvements in root systems for optimal climate resilience.