2023-12-21 シンガポール国立大学(NUS)
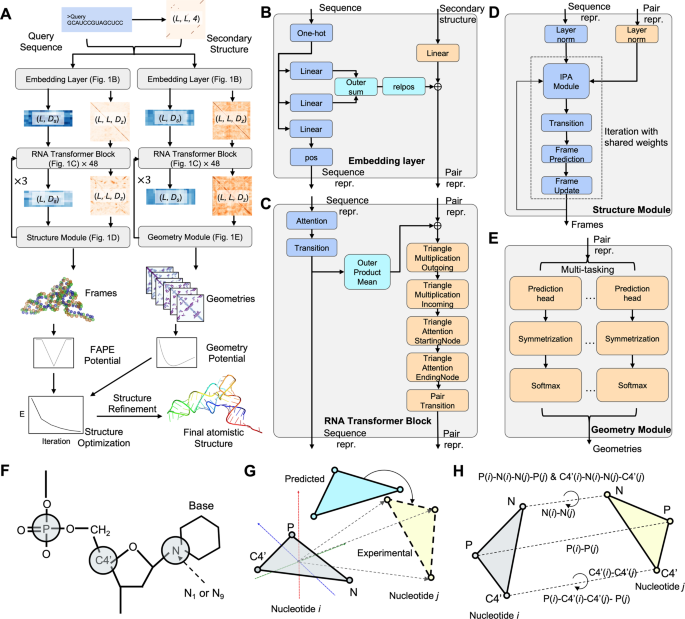
◆この方法は、伝統的な手法に比べてRNAモデルの精度を70%以上向上させ、次世代バッテリーの進化や新薬の設計に寄与する可能性があります。DRfoldはAI力場の精度を向上させ、RNAの三次構造を予測するために2つの深層学習ネットワークを組み合わせています。
◆この新しいアプローチは、RNAの予測において従来の方法よりも優れた精度をもたらし、RNA分子の生物学的機能の解明や新しい薬物の開発に寄与する可能性があります。 DRfoldはオープンソースであり、他のRNA関連の問題の解決にも適用可能です。研究チームは今後、AI戦略をプロテイン-RNA相互作用に拡張し、DRfoldの精度向上を目指しています。
<関連情報>
- https://news.nus.edu.sg/ai-based-approach-rna-3d-structure-prediction/
- https://www.nature.com/articles/s41467-023-41303-9
第一原理RNA構造予測に深層幾何ポテンシャルとエンド・ツー・エンド学習を統合 Integrating end-to-end learning with deep geometrical potentials for ab initio RNA structure prediction
Yang Li,Chengxin Zhang,Chenjie Feng,Robin Pearce,P. Lydia Freddolino & Yang Zhang
Nature Communications Published:16 September 2023
DOI:https://doi.org/10.1038/s41467-023-41303-9
Abstract
RNAs are fundamental in living cells and perform critical functions determined by their tertiary architectures. However, accurate modeling of 3D RNA structure remains a challenging problem. We present a novel method, DRfold, to predict RNA tertiary structures by simultaneous learning of local frame rotations and geometric restraints from experimentally solved RNA structures, where the learned knowledge is converted into a hybrid energy potential to guide RNA structure assembly. The method significantly outperforms previous approaches by >73.3% in TM-score on a sequence-nonredundant dataset containing recently released structures. Detailed analyses showed that the major contribution to the improvements arise from the deep end-to-end learning supervised with the atom coordinates and the composite energy function integrating complementary information from geometry restraints and end-to-end learning models. The open-source DRfold program with fast training protocol allows large-scale application of high-resolution RNA structure modeling and can be further improved with future expansion of RNA structure databases.