2023-09-11 カリフォルニア工科大学(Caltech)
◆遺伝子発現データを正確に特定することは、パーソナライズ医療や疾患研究など多くの分野で重要です。この研究では、scRNA-seqの手法に関連する問題が特定され、参照トランスクリプトームの最適化が遺伝子発現情報の損失を防ぐのに役立つことが示されました。最適化された参照トランスクリプトームは、細胞タイプや状態の新たな発見に寄与する可能性があります。
<関連情報>
- https://www.caltech.edu/about/news/invisible-cell-types-and-gene-expression-revealed-with-sequencing-data-analysis-improvement
- https://www.nature.com/articles/s41592-023-02003-w
RNAシーケンスの欠損データを最適化されたトランスクリプトームリファレンスで回復させる Recovery of missing single-cell RNA-sequencing data with optimized transcriptomic references
Allan-Hermann ,Helen Poldsam,Sisi Chen,Matt Thomson & Yuki Oka
Nature Methods Published:11 September 2023
DOI:https://doi.org/10.1038/s41592-023-02003-w
Abstract
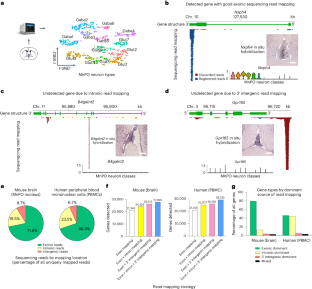
Single-cell RNA-sequencing (scRNA-seq) is an indispensable tool for characterizing cellular diversity and generating hypotheses throughout biology. Droplet-based scRNA-seq datasets often lack expression data for genes that can be detected with other methods. Here we show that the observed sensitivity deficits stem from three sources: (1) poor annotation of 3′ gene ends; (2) issues with intronic read incorporation; and (3) gene overlap-derived read loss. We show that missing gene expression data can be recovered by optimizing the reference transcriptome for scRNA-seq through recovering false intergenic reads, implementing a hybrid pre-mRNA mapping strategy and resolving gene overlaps. We demonstrate, with a diverse collection of mouse and human tissue data, that reference optimization can substantially improve cellular profiling resolution and reveal missing cell types and marker genes. Our findings argue that transcriptomic references need to be optimized for scRNA-seq analysis and warrant a reanalysis of previously published datasets and cell atlases.