2025-10-09 東京大学
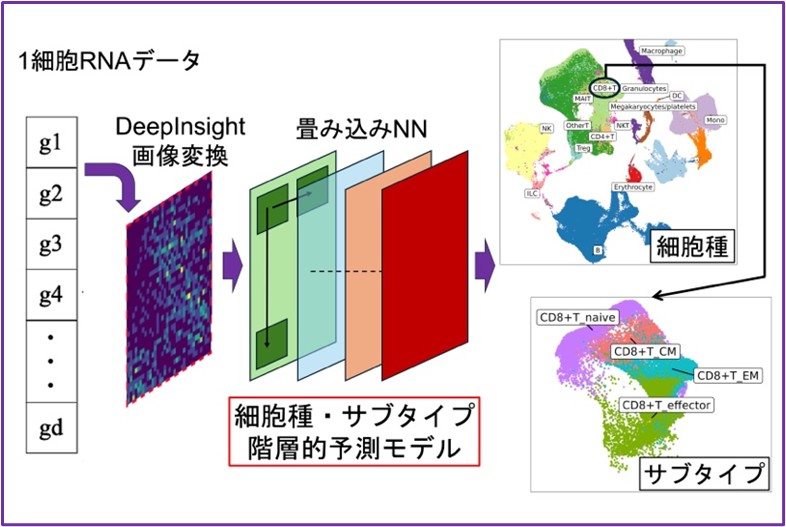
提案手法scHDeepInsightの概略図
<関連情報>
- https://www.s.u-tokyo.ac.jp/ja/press/10937/
- https://academic.oup.com/bib/article/26/5/bbaf523/8276461
scHDeepInsight: 単一細胞RNA-seqデータにおける正確な免疫細胞アノテーションのための階層型ディープラーニングフレームワーク scHDeepInsight: a hierarchical deep learning framework for precise immune cell annotation in single-cell RNA-seq data
Shangru Jia, Artem Lysenko, Keith A Boroevich, Alok Sharma, Tatsuhiko Tsunoda
Briefings in Bioinformatics Published:09 October 2025
DOI:https://doi.org/10.1093/bib/bbaf523
Abstract
Accurate classification of immune cells is crucial for elucidating their diverse roles in health and disease. However, this task remains very challenging in single-cell RNA sequencing (scRNA-seq) data due to the complex and hierarchical relationships of immune cell types. To address this, we introduce scHDeepInsight, a deep learning framework that extends our previous scDeepInsight model by integrating a biologically-informed classification architecture with an adaptive hierarchical focal loss (AHFL). The framework builds on our established method of converting gene expression data into two-dimensional structured images, enabling convolutional neural networks to effectively capture both global and fine-grained transcriptomic features. This design utilizes hierarchical relationships among immune cell types to enhance the classification ability beyond the flat classification approaches. scHDeepInsight dynamically adjusts loss contributions to balance performance across the hierarchy levels. Comprehensive benchmarking across seven diverse tissue datasets shows scHDeepInsight achieves an average accuracy of 93.2%, surpassing contemporary methods by 5.1 percentage points. The model successfully distinguishes 50 distinct immune cell subtypes with high accuracy, demonstrating proficiency for identifying rare and closely related cell subtypes. Additionally, SHAP-based interpretability quantifies individual gene contributions to reveal the biological basis of classification decisions. These qualities make scHDeepInsight a robust tool for high-resolution cell subtype characterization, well-suited for detailed profiling in immunological studies and extensible to nonimmune cell types.